-
Donor Model Evaluation
- Best model(s) according to ROC measure:
C=10.0 shift=0
Best model(s) according to PRC measure:
C=10.0 shift=2
Best model(s) according to accuracy measure:
C=10.0 shift=2
Detailed results:
C shift ROC PRC Accuracy (at threshold 0)
0.1 0 99.0% 88.1% 96.3%
0.1 1 98.8% 87.1% 96.3%
0.1 2 98.8% 87.5% 96.3%
1.0 0 99.0% 88.2% 97.5%
1.0 1 98.8% 87.4% 97.7%
1.0 2 98.8% 87.8% 97.8%
10.0 0 +99.1% 87.9% 98.4%
10.0 1 99.0% 87.5% 98.4%
10.0 2 99.1% +88.6% +98.5%
SVM Cross-validation with Spectrum Kernel
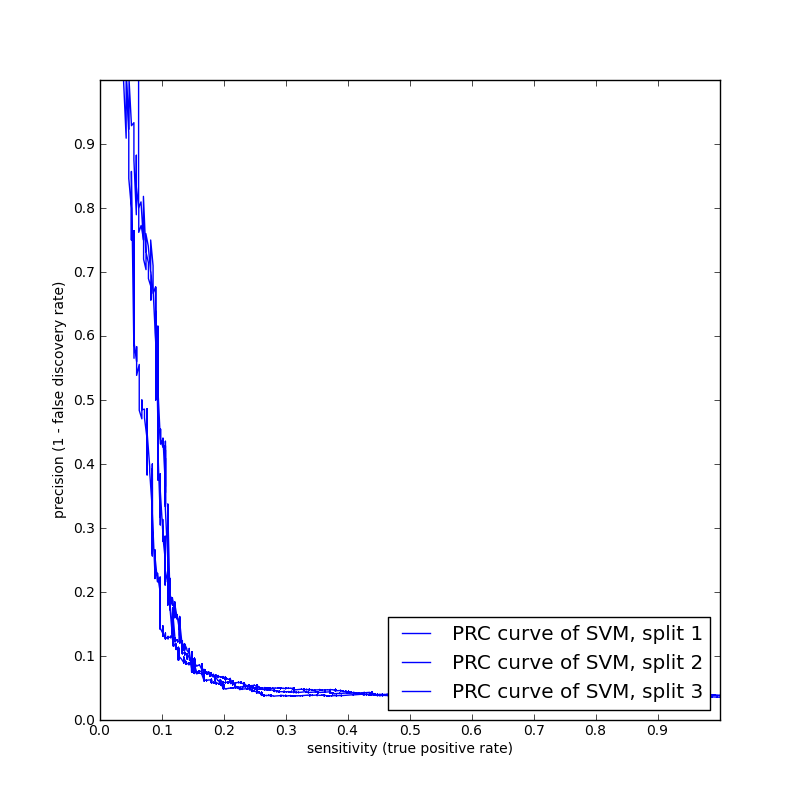
We performed some cross-validation benchmark runs using the basic Spectrum Kernel. Here are the auPRC curves obtained foracceptor and donor on sampled data sets of 20K worm sequences. The spectrum kernel did not do very well, as it is basically composition-based, and lacks information available in positional, regional, correlational, and other more complex features.
 |
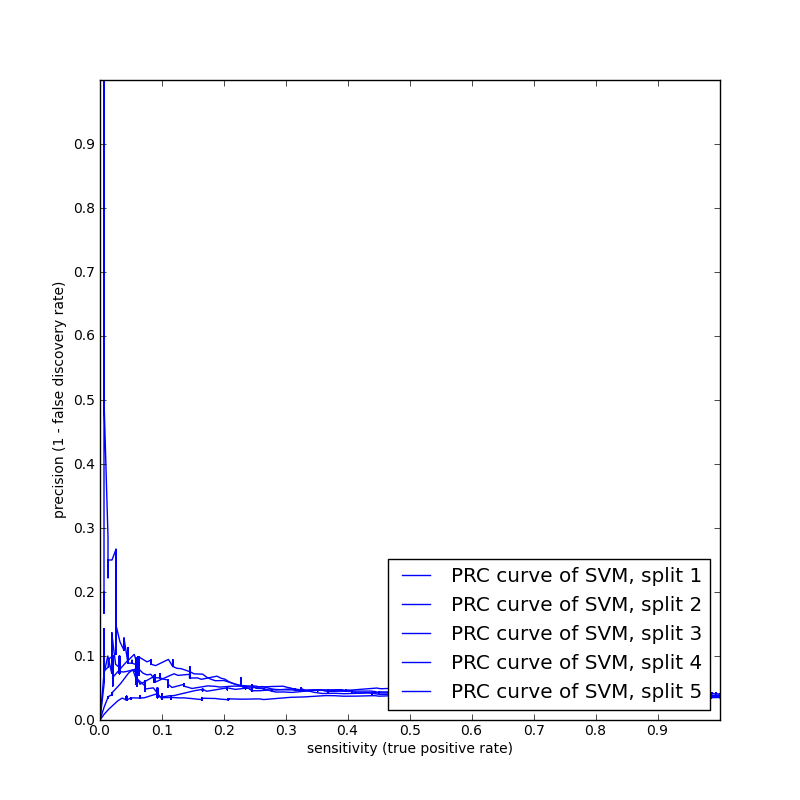 |
| Acceptor 20K run with 3 fold
cross validation |
Donor 20K run with 5 fold cross
validation |
SVM LibLinear Model for FG-EA
- SVM Training libsvm tested with B2Hum (Since LibSVM files
are large, they can be provided upon request)
- Acceptor
- Donor
- Top Features for Human trained and tested with B2Hum
- SVM Training cross validation with Worm (Since LibSVM files are large, they can be provided upon request)
- Acceptor
- Donor
- Top Features for Worm (C_Elegans)
Gene Annotation for 5 Sequences (Scores at all positions, Exon markings, TP/FP data)
We took 5 Random Sequences of B2Hum Genesplicer sequence and for each window (162 nt same as training window) starting from position till end, predict position +80 (83) for donor and acceptorpositions. We didn't include the first 80 and last 80 as they will be outlier due to filler random positions.
Sequence 1:
Sequence Data:
| tgtgggccaaagctgaccaactcccccaccgtcatcgtcatggtgggcctccccgcccgggg caagacctacatctccaagaagctgactcgctacctcaactggattggcgtccccacaaaag gtgagactgggtctcgaggccggacccctgctcgtgcaaaaacttgaccttccatctcagtt gctggtttcacaggcaggaccacccctgggcgcctctgtcccctggtcggggagtctgatcc tgactcccatgcagctgggtccccttcacactgtgtctacaccctttttcttctgttggggt tttctcctcactggccattctctttcccactcccctaagtcctgcccgactcagatttcacc tccttcaaagccatctccagccacctcagtcctttacattcctgtcgcactcagtcctagct aacattcctctgctgatagccctaagcactgcctaccgtagatcaagggtcttcaaccccca ggctgtggactgctatgttgggtggcctgttaggaacctggctgcaggaggtgagtggcagt gagtgcgcattcctgcctgagctccgcctccagtctgatcagtggtggcattagattcccat gggagtgaaccctattgtgaactgggcatgtgatggatctaggttgtgtgctccttatgata atctaatgcctgatgatctgaggtggaacagtttcatcccaaagccgtacacccacaacctc cctgtccgtggcaaaattgtcttccacgaaagtggtccctggtgccaaaaaggatggggact gctgttgtggatgacacatttaagaatctctgcttggtcttcccaactagatggggaacatc ttgtggataggcactgggtattttttttttttcaattaattttattttagattcgggtacat gtgcatgtttgttacctgagtatattgcatattgattggggattgggcttctagtgaacctg ttacccagacagtgaacattgtacccaactggttattttttaacccttgctcatctcccacc ctcccctcttttgaaaggatggtttttatgtttaccatccccgctgtgcccagctcagtgcc caggaaccatgggacacatgtctccggccctggccctggcggctgcaccagcagaagggttg acctagatctggcggtgggcatgaggccccactcgctttctgataccattggtggaaatgtg ttgtccctatttttgctagacttggggcctccccccacccatcctcacagtctcagtctctg gtttaggatattgtgggactgagattgtagacttggctggaggggacagtgtcgcagagcat gcggcaggggcattgcagctggactggggaagccgggcagcctgtgccctgctgtcctcagg agttgagggtagcgtaggactgggcgccggcattgcttccacaccatctcattcaagtcggt gtggttggcctggtggctcttcctttggtcgatcctatggtcccggtgtgagctggcccctt cctcctgctcgatcatccagactgttctctttcccgtccacagtgttcaacgtcggggagta tcgccgggaggctgtgaagcagtacagctcctacaacttcttccgccccgacaatgaggaag ccatgaaagtccggaagtaaggctgggccgcgggcgtagggctgggctgtgggaataaggct gggccgcgggcataaggctgggctgcaggagtaaggctgggccgcgggcgtagggctgggct gtgggaataaggctgggctgcggggctgcgggtgtaaggctgggctgcgggcttgggcttgc tttgttctgagccaggctctgtaggaggcagagaaagccggcctgcgggtgggtcctgctgg tggcttcagggctgtgcagtgtgggaacaaggttgctgctctcatctgttccctgagggtct cgctgagctgaccctcactgggacagagccgcagaagcaccctcactgagaattagcaagtc ctgttcatcgggagtgtctgtttatgtttggaaaggatttcttgttaagtgcgagttgattg tgaaaccctagtgcaagggtgttcggccccggctgtatgctggagtctcctgggggatgcta aaatccctgatgccaggccccaccccagaccagttagtgcaactctgtgggtggctctgggc agccatttcttagcagctccccaggagtggttcccgcgtgcagcccggtgcgagcaccgcct cccgagggttcctgaggccacccgtgtggggccccaggttggaagcctctgggcctgagggg tgctggccagcctgcccagcgctaagcagtgtagaatggaaccaccagttactctgcttgtc ggggcgcgtcgggagttgcttgggctgccccagcggatgcaccagcgctgacattcgggcaa tgtcttgtgcattccaggcaatgtgccttagctgccttgagagatgtcaaaagctacctggc gaaagaagggggacaaattgcggtaagtccaggcaatgtagccggctcggtggtccagtccc acccatgagggttgtcctcaccggcctgggttgtacccgagaccctgtgcctctgcccctga gggtggaggaaagcaggtgccaggccagcccaatggctcaggaaggcggatgtggggcggac tcagggacccaccccctctgtggccagtcccacacctccagggcgggcaggactcgagttac ccacaggaccactggcccttccttgtacactgtcccattgcacattactgttttggttgttt gcttttggggtatttcccccacgctaccacccctgagtgctgttggtagttgcgggaggcct gcctcctaagcactccctggagaggactgtgggcaggcacaagctcccctgaagcacttttg gggcttattcctggagatgatgggtctggcatctttgctgttctggtaaacgttgttaaact caaacttagagttttcttcctggccctttgctctccatatcaggttttcgatgccaccaata ctactagagagaggagacacatgatccttcattttgccaaagaaaatgactttaaggtgagc tgagcgtcttgtgttgtgctagagggctcgggagagaggaaaaggcctcaggaagcggcctg agtcctgcttggtccttcccctcctgctgccatgttccttagtcctgctcggtccctcccct cctgctgcccacgttttgtggccttaaatttgggcctttgtgtggtagggctcttgggggcc gtggggcgttccagaggctgcagtgaggaggtgggctttcctgtgtttgggagttggtgggt ggcgtgcttccgccttgtccgggtgatgacatcgcagtgatggcaaggttgatccttcgtgg tgtttgttttctaacatccccacctcacttccaccaggcgtttttcatcgagtcggtgtgcg acgaccctacagttgtggcctccaatatcatggtaagacagccgggagccccgtgcttctgc ggcagcgtagaccacaagggcatttgcagtctgaggaagtgggggaccgggtccagctcgga agcgcagctgagcccttag |
Exons
45437_AB012229 1 124
45437_AB012229 1594 1690
45437_AB012229 2498 2564
45437_AB012229 3082 3156
45437_AB012229 3510 3677
Predictions:
1. Showing Top line actual exons start-end
2. SVM scaled score [0,1] for each position of window length 162.
Observations:
1. Exon2, Exon3, Exon4 rightly identified
2. Initial Exon acceptor not considered due to boundary condition (first 80 are considered outliers)
3. Final Exon donor not considered dur to boundary condition.
4. One False Positive Donor at 80 corresponds to GT.
Sequence 2:
Sequence Data:
| atggtaggtgtttcgtttcttgcttctttttccttgccggcggagacccctaagctgtattc ccattgcccctagtcatccactccctaccatggtcgggggttccaggctgcgcatggccgcc tgcggggcagggtggccggcgcgggcccggggcggggctcccggagccgtgtgttaggcccg cggttcggatctctaggacacgcgggcccctgcgctaccgtggtgagacctcacggccctga gcggatcggtaccctcagctttcccaaacgctccagaagttaggtctttgacccacaggctt acaggaccatctcggctggcgggcatcgccccctgcccctaattccttaggccttaccacca agctttttccacacagccatccagactgaggaagacccggaaacttaggggccacgtgagcc acggccacggccgcataggtaagtgccggcttcccctcggggtgggccttgggctctcttcg ggtgcttagctagtctggagatcggtagcctataagtgggttagaataagacctttttgtgg tcaagttgcacagctgttgatttttttctgacgatcctctagtattccagttctaaggaatt tcacatcagtggggtaataggaattgagcaggcacggtattgggttagttgaagacatggag tactgtgggaatgctgtgatgtggaacctgaaaagatgtttcacccggaatcctaaagtaat cgcattgctgaaaaccggcatcggtaggtgggaacagcgtaagcgggacacagaagtctggg aaacactctgcttttgtgcgaagaagtattgagatgcatgaagaagctgtgtggtgcatgta gcttttttgtgtgtgtgtgagactgatcactgtcgcccaggctggagtgcagtggcgaaatc tcggctcactgcaacccctgcctgccgggctcaagcgattctcctgcctcagcctcccgagt agctgatattacaggtgtgcgccacgacgcccggctaattctttttctatttttagtagagt cgggggtttctccgtgttggccaggctggtctcgaactcctgacctcaggtgatctaccccc cctcgtcctcctaaaatgctgggattacaggcatgagccatcacaccggccccacgtagctt tgtattcctgcaggcaagcaccggaagcaccccggcggccgcggtaatgctggtggtctgca tcaccaccggatcaacttcgacaaatagtaagtgtccttggactgcttttattgaaacagct tgggaggtaggggcagagagagggctggcttaaacaaaaagtttagaagcaagccttgccta ttgctgttttttaccaagttaacacttggtgtgaactgagaacctgtcatcgaggctagagt cacgcttgggtatcggctattgcctgagtgtgctagagtcctcgaagagtaactgctgacct tattcactggctgtgggccttatggcacagtcagtcaccaggttagagacatgcttcacatt cacctacccacaaactagtggatgataaattttggctattcagaagacgtttattataggag tatgtagattttccatagagtgctgttatgtgacttgaattttagtctcggccctgcctctg acattgtcggtggtttatcctggttccaggaaataagactagccttttcctcatgatagtct ttggtggtttttaaaacagttgtttaagtcaacagatgtatcatatgcctgacactgctcta caccagtgaataatttacactctaatagggggtggtaactataaagatgataaacatagcat cttaattggagtgtgtatgaaggtggttgttacctcttcctagccacccaggctactttggg aaagttggtatgaagcattaccacttaaagaggaaccagagcttctgcccaactgtcaacct tgacaaattgtggactttggtcagtgaacagacacgggtgaatgctgctaaaaacaagactg gggctgctcccatcattgatgtggtgcgatcggtaagttaattggatgtttttctgtacttc cataccttcccttacaaaactctggcttaatctaatccacttatataatctgtacttcccag ttacctaccagacattgatattcttcctgtggtagaattatcataggtagttccctatccgt agcagtgcctactgtcactgcccaggttgtatcaggtttgcatttcgtgcttgaactatagc tggttttcactgagcacagctcttggcccttcatgttctccagataatagaatcctaatatg ttccattgatactcagtgccatgcattatctgaagagattttcccccaaaacagatgtatta tgtctgtccttgcgggggttctggtccctgtgtcagtcttaactctcatgaatatagaggta gtgttaagaggccagaacctagggacgctttaaattcacttcccagcctatttaatgtccat tgagtagttctggtggtcaggaaggtggttgtcttcttttgcttagcagggggtatttgagc aggaggaggcttatgctttgccgagactagagtcacatcctgacacaactcttgtcctggtg tgctagagtactcgaagagaatctactggtcttgattcactggtgggggcagtcggtgcccc cgttagtgcccagatcagaaacatacataccctgcctagggatttagaaagtgggttggcag tctttcctcacgcccatcacgcagttggtacctactacagtgtattgtaaacttttttctct gttcttctagggctactacaaagttctgggaaagggaaagctcccaaagcagcctgtcatcg tgaaggccaaattcttcagcagaagagctgaggagaagattaagagtgttgggggggcctgt gtcctggtggcttga |
Exons
45514_AB020236 1 3
45514_AB020236 389 452
45514_AB020236 1192 1267
45514_AB020236 1904 2078
45514_AB020236 2863 2991
Predictions:
1. Showing Top line actual exons start-end
2. SVM scaled score [0,1] for each position of window length 162.
Observations:
1. Exon2, Exon3, Exon4 rightly identified
2. Initial Exon acceptor nd donor not considered due to boundary condition (first 80 are considered outliers)
3. Final Exon donor not considered dur to boundary condition.
4. One False Positive Donor at 80 corresponds to GT.
Sequence 3:
Sequence Data:
| atgctgctcctgttcctcctcttcgagggtctctgctgtcctggggaaaatacagcaggtaa gaagagtgcaggtggaaagatacctatggtagggcaccagagggctgagaggaagctctggg gaggtcctgggggagggagcagtactcttctaggatgcccttggaatatgcctttcaggcta gttccaggcagagaattcttgctctcagtctcagtttttgtctctgattttggagaaaggaa gctggccccacaggaaaagggtattggagtatgtacaagctacctaactgtctctcatctct gggttccttttttccctttggcatcacttttcccatccctttacattctctctacttgtcat ttccctctctctcagctccccaggctctacaatcctatcatctagcagcagaggagcagctg tccttccgcatgctccaaacttcctcctttgccaaccacagctgggcacacagtgagggctc aggatggctgggtgacctgcagactcatggctgggacactgtcttgggcaccatccgctttc tgaagccctggtcccatggaaacttcagcaagcaggagctgaaaaacttacagtcactgttc cagttatacttccatagttttatccagatagtgcaagcttctgctggtcaatttcagcttga atgtaagttcgttgctctaagctgataatttgcctgggaacaccaactatttccaaatgaag atagatatatagactctgaccatcatttaaccttactaaccttgttccccactctctgactc ccactccctcctctgcttcacccttcaccaccacccacactccaccatatacacaaaagggc ctgcatgtacatatctcaacatgaatatagcttcatgtctggctctttggaatgattgtctc ctctggatcttctgcccctcattcctgccctcagactcagcctttctcaaccctctttctgc ccttctttatcctttgcctgagtgttgacatggactggcctgtacctaaccactttcacgtg aattatttatgaccaatctcctatcttcctgatagccttcccatctaccactttcccattag ttatttcaaagtatctttattatcttcaaattttcttcccacaaattttcttcccctttgcc agtaaactctagtctccatatgatttcccagcaaatttttcttcccttgaatctctttactg tctaaattgtttgtttttcttccttgtcattctttccataatgatctctcttccctgtccac tctcagaccccttcgagatccagatattagctggctgtagaatgaatgccccacaaatcttc ttaaatatggcatatcaagggtcagatttcctgagtttccaaggaatttcctgggagccatc tccaggagcagggatccgggcccagaacatctgtaaagtgctcaatcgctacctagatatta aggaaatactgcaaagccttcttggtcacacctgccctcgatttctagcggggctcatggaa gcaggggagtcagaactgaaacggaaaggtgagcccaactctctctctcccctcttgttcct agtactataactctcatatttgaatttgcctctcatcatcattttgaaagacatagtgagag actagagaatgagatgtgtgggttcaggactgtttcttagacaagagaaagaagtgattact aaatcactcttagtattattacaaaggcacctgagtctctgagctctggcctggggtgccct tcaaaattccattttttttctatcttcttcttcctagtgaagccagaggcctggctgtcctg tggccccagtcctggccctggccgtctgcagcttgtgtgccatgtctcaggattctacccaa agcccgtgtgggtgatgtggatgcggggtgagcaggagcagcggggcactcagcgaggggac gtcctgcctaatgctgacgagacatggtatctccgagcaaccctggatgtggcggctgggga ggcagctggcctgtcctgtcgggtgaaacacagcagtctagggggccatgatctaatcatcc attggggtgagaaacagctgaggctctgctgggaaataatgaaaatagccctggggcttttg agtgtggggctgaggaaatgggtaggaatgctaggtacaagaagggtaaaactgggacaatc aaaataaagaaggatagagtatgacagtagttaaattttaagaaaatggaagtagagaatta gacatactaacagaaaaaggaggaggaactagtgatttagtgggagagggttgggaggagat cacagacaaaggatcaggaggaattgaaatgagggctttggaaaacccagatgaaaattcta ggaaggtcccacccttgtgaaatgggaaatctcagcttggtggaatagagtattttagggtt ggtattcttattctatccccaaccaggtggatattccatctttctcatcctgatctgtttga ctgtgatagttaccctggtcatattggttgtagttgactcacggttaaaaaaacagaggtga |
Exons
45868_HSCDIR2 1 58
45868_HSCDIR2 418 684
45868_HSCDIR2 1309 1578
45868_HSCDIR2 1836 2114
45868_HSCDIR2 2507 2604
Predictions:
1. Showing Top line actual exons start-end
2. SVM scaled score [0,1] for each position of window length 162.
Observations:
1. Exon2, Exon3, Exon4 rightly identified
2. Initial Exon acceptor and donor not considered due to boundary condition (first 80 are considered outliers)
3. Final Exon donor not considered dur to boundary condition.
4. One False Positive Donor at 81 corresponds to GT and one false positive acceptor around 80 .
Sequence 4:
Sequence Data:
| ttcattccacagacacacacagcctctctgcccacctctgcttcctctaggaacacaggtaa gagcttcaagcctctccagcttaataacatgaattatttttgagaataataatgatactgtg ttctatatcatgcatctcctgcattctgtctgattatattttacttattctgccagagcaaa attaaaatacctatttcatctgatttgtcctttatctaaattgcttagttccaagtaaacca aggcacttttagaacacagagggagagtgccttgcagccagagagtcttgaaggagatgtca gggacgcatcttaacagctggttggatgtgatccacagaggtctcctgttagcattcattgt aaagccatcctacctagctctagtgtaaccagcaatgaaagaaagataaagaaagataaaga gggtcgattacttatttacaatagtctttaaaaacgtagttttgtaagccttctaattagga cattaatatatttaatatatgcacattgtagaaagattgaagcgttaaaaataagagaaaaa ctttaaatgtcaaaatctcacaacccagatatatcatttctttaagaaaattgtactacaaa ataccattccatttattaaagtcattctgacaggaatctgatgcttttccaggagttccaga tcacatcgagttcaccatgaattcactcagtgaagccaacaccaagttcatgttcgatctgt tccaacagttcagaaaatcaaaagagaacaacatcttctattcccctatcagcatcacatca gcattagggatggtcctcttaggagccaaagacaacactgcacaacaaattagcaaggtagc tatcagcatcattacgttgtcctgttgcagtttttctctggttccgtcggctagcacgcaga tggtaatagatgtggtggtctgatgggtagcacagggggctgtgcaggaattcccataactg tgagaccactgacttaaacagatcttttgagtaaagttttcttgtcccgcttcatgtctctt ccaggttcttcactttgatcaagtcacagagaacaccacagaaaaagctgcaacatatcat |
Exons
45378_AB005548 1 58
45378_AB005548 673 863
45378_AB005548 1059 1115
Predictions:
1. Showing Top line actual exons start-end
2. SVM scaled score [0,1] for each position of window length 162.
Observations:
1. Exon2 rightly identified
2. Initial Exon acceptor and donor not considered due to boundary condition (first 80 are considered outliers)
3. Final Exon acceptor and donor not considered dur to boundary condition.
4. No False positives in acceptor and donors .
Sequence 5:
Sequence Data:
| atgcttgcgggtgccgggaggcctggcctcccccagggccgccacctctgctggttgctctgt gctttcaccttaaagctctggtaaggagagtaaggggagggaaaagagtcactggaggacaga ttcgttccccggcctccgggctggggagattcttatccatggacagcgcaggagggggtgccc tattttctgggttttgacttttgtcccttcccccaactccgctacagccaagcagaggctccc gtgcaggaagagaagctgtcaggtgggtgattgtctgggaacccgacacgtcccgtcgtttgt ttttggttttagtatcttccccatataccaagagcacaggtggaaaaggccggcaccacatgg aatgagggcaagcgctgtgccgggaagttggatgggcagagcctgggagaaatgttttctgaa tggcctggggctggcatttatctttcctttcagcaagcacctcaaatttgccatgctggctgg tggaagagtttgtggtagcagaagagtgctctccatgctctaatttccgggctgtgagtatct atttctctaccctttttttttttaaacatctaagcaaacacttaagcaacatcttaacatcca agtcttattaagctgcctttgcatcatctttttttctttcttcttgcagtctcagtgtatgtc caaatctgtacttgtctttcccctgaaaatgtgcccctttctttccaagctaatggcagtatt caactgataactatggtttttttgtttgtttttgagacggagtctcactctgttgcccaggct ggagtgcagtggcgagatctcggctcaccacaacctccgcctcccgggttcaagcgattctcc tgcctcagcctcctgagtagctgtgactaaaggcgcgttgcaccatgcctggctaatttttgt atttttagtagactcgggtttcactgtgttggccaggctagtctctaactccttacctcgtga tctgcccgcctcagcctcccaaagtgctgggattataggcgtgagccgccacacccggcctca aataactatgttttattcacttttagtatagtaggctctggaatggaatgtatctttgccact cctagactgttgcccctgaagtgttctaacatacattcgtaatcatgcaaccaccacctccac catccgcatcagaactctttcattagctctctgttgcctaccactccaaaccatagcagttgg gcacctgcaccttctgaatggcagcctttttgtttatcctgttgccccttcctaacatgtact ttgctccttttctcctggcagaaaactacccctgagtgtggtcccacaggatatgtagagaaa atcacatgcagctcatctaagagaaatgagttcaaaaggtgagtgctgtttctcttctctgag atcacctcctttgctcctatgactttagatggatggtcccaccttttcccctggtctgggggt agcatggaatatctgaaagaagagggggccgggctcacacctataatcccagtacttagggag gctgaggctaacagatcacctgaggtcaggagttcgagatcagcctggccaacgtggtgaaac cccatctctactaaaaatacaaaaattagctggtgtggtgggtgcccataatcccagctactt gggaggctgaggcagggagaatcacttgaacccaggaggcagaggttagagtgagccaagatc atgccactgcactccagcctgggcaatggagcgagactgtgtgtcaaaaaaaaaaaaaaagag ctgggagggtggctcacgcctgtaatcccagcactttgggagccgagcggtggatcacgagtt aggagttcaagaccagcctgccaagatggtgaaactcatctctcttaaagatacaaaaattag ctgggcatgatggcgggcacctgtaatcccagctacttgggaggctgaagcagaaaactgctt aaacccgggaggtggaggttgcagtgagccaaggtcacgccactgcactccagtctgggtgac agagcgagacaccatctcaaaaataataagaaagaagacgggatttggagccaaattacttga acccaagtcccagctctactcctcagcactatgttattggccatattttctcatcagtaaaat tggttaatactagctctgcttatccagagaataaattaaatgatctatttgagagggatttat gtactatacacacaatagaagcctggctctgatttcctcttgagtgtatcctggctaggctga agtagaagggaaattagagggagaagctgtctggccttccttgggaataactattccttcctg ccctcagctgccgctcagctttgatggaacaacgcttattttggaagttcgaaggggctgtcg tgtgtgtggccctgatcttcgcttgtcttgtcatcattcgtcagcgacaattggacagaaagg ctctggaaaaggtccggaagcaaatcgagtccatatag |
Exons
45463_AB016492 1 83
45463_AB016492 237 274
45463_AB016492 475 557
45463_AB016492 1345 1424
45463_AB016492 2402 2558
Predictions:
1. Showing Top line actual exons start-end
2. SVM scaled score [0,1] for each position of window length 162.
Observations:
1. Exon2,Exon3, Exon4 rightly identified
2. Initial Exon acceptor not considered due to boundary condition (first 80 are considered outliers)
3. Final Exon donor not considered dur to boundary condition.
4. One False positive in Donor 82 position .
Training, Testing, Interpretation Times
Some recorded times are provided below. We employed a Windows XP 16GB machine with 4 cores, Windows XP.| Note | Fasta Files Size | Sparse LibSVM/Liblinear files | Evolutionary Run Time | Feature Interpretation time | SVM Cross Validation Time |
| Acceptor/Donor (20K) | 2.8 MB | 563 MB | 2.5 Hours | 1 hour | 15
minutes
(LibSVM) 2 minutes (LibLinear) |
| Acceptor/Donor (180K) | 25.68 MB | 8 GB | 6 Hours | 2.5 hours | 5 minutes (LibLinear) |