One of the research thrusts in the Shehu lab is understanding the modulation of biological activity in protein molecules via a process known as conformational switching. One of the ways we do so is by designing powerful, sample-based algorithms to extract a discrete representation of the conformational space of a protein and additionally model switching between conformations of interest.
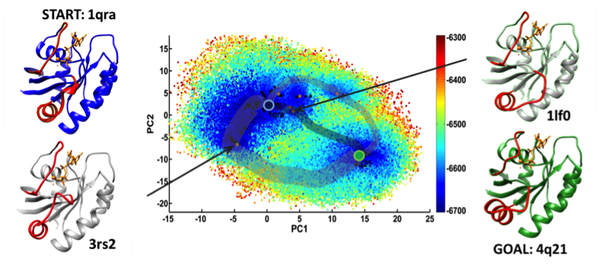
The image shown above synthesizes findings of a robotics-inspired algorithm designed in our lab, published in IEEE TCBB 2016, for a hallmark protein of critical importance to human biology and health, H-Ras. Specifically, the image shows the explored conformational space embedded on two collective coordinates and the transition tubes that summarize all possible conformational switching routes between two conformations of interest. Specific hypotheses regarding possible mediation by other experimentally-known conformations can also be investigated via the computation of tours.
The algorithm leverages experimentally-known conformations of healthy and disease-related variants of a protein to obtain an ensemble of relevant conformations within a reasonable computational budget. The ensemble is then embedded in a nearest-neighbor graph, which is queried to obtain the lowest-cost path and other similarly-costly paths connecting any two conformations of interest. Extensive analysis on H-Ras, its oncogenic variants, and other proteins suggests the algorithm is informative and can be used to model structural excursions that are challenging to directly simulate via physics-based simulations of molecular dynamics.
This work is supported by grants from the National Science Foundation (Grant Nos. 1440581 and 1144106). For more information, visit the Shehu lab.
